If you could collect all the organisms from the ocean surface down to 200 meters, you would find that SAR11 bacteria, also known as Pelagibacterales, would account for a fifth of the total biomass. Though these tiny organisms are invisible to the naked eye, their role in global nutrient cycles is significant.
SAR11 bacteria have adapted to thrive in nutrient-poor marine environments, influencing processes that are crucial for the planet’s ecosystem. Despite their importance, the precise mechanisms through which these bacteria impact nutrient flows were unclear – until now.
A recent study published in Nature by researchers from the Okinawa Institute of Science and Technology (OIST) has shed new light on SAR11’s role.
Dr. Ben Clifton, the first author of the study, explains: “We knew that SAR11 is a key player in important nutrient cycles, such as carbon and sulfur exchanges, but we didn’t know the full extent. Now, by comprehensively mapping out the transport proteins of the bacteria, we have a much better picture of how SAR11 slots into these cycles.”
Professor Paola Laurino, the senior author, attributes this breakthrough to global seawater sampling initiatives like the Tara Oceans project, saying: “These datasets have allowed us to link genomic data to protein function.”
Decoding SAR11’s transport proteins
Transport proteins, which facilitate the movement of nutrients into and out of bacterial cells, are critical to how bacteria interact with their environment. For SAR11 bacteria, which are ubiquitous in the oceans, nutrient uptake has a broad influence on global cycles. However, studying these bacteria is challenging due to their specific growth conditions. To address this, the researchers genetically modified E. coli bacteria to express SAR11 transport proteins, enabling them to examine the proteins in the lab.
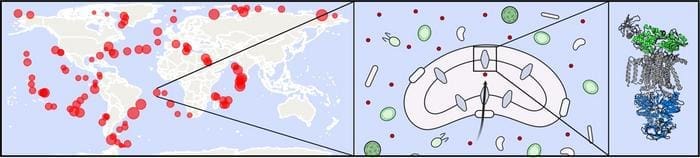
By analyzing genes across the SAR11 metagenome – the combined genetic material of all SAR11 species – the researchers identified genes linked to essential marine processes. One of their key findings was a protein that interacts with dimethylsulfoniopropionate (DMSP), a compound crucial to the sulfur cycle and climate regulation.
In total, the team discovered thirteen transport proteins, responsible for handling compounds like DMSP, amino acids, glucose, and taurine, all of which play significant environmental roles.
Shaping global nutrient flows
“Through these experiments, we discovered specific properties of transport proteins that enable SAR11 bacteria to thrive in nutrient-poor environments. This could not have been discovered from just looking at the genomic makeup alone,” Dr. Clifton noted.
But the research on SAR11 is far from complete. Now that they have identified the proteins involved in nutrient transport, the team is exploring the metabolic pathways that determine how these nutrients are used and converted within the bacteria. Furthermore, in collaboration with the Weizmann Institute of Science, the researchers are investigating how SAR11 interacts with its surrounding environment before absorbing nutrients.
This research is part of a broader trend that connects environmental DNA to protein function, opening up new opportunities to understand how microscopic life shapes global processes.
“By bridging the macro perspective of marine diversity and the micro perspective of protein biochemistry, we’re setting the stage for further questions about how bacterial proteins fit into global nutrient cycles, and how these bacteria contribute to, and are affected by, climate change and shifts in ocean biodiversity,” says Prof. Laurino.
Journal Reference:
Clifton, B.E., Alcolombri, U., Uechi, GI., Jackson C.J., Laurino P., ‘The ultra-high affinity transport proteins of ubiquitous marine bacteria’, Nature (2024). DOI: 10.1038/s41586-024-07924-w
Article Source:
Press Release/Material by Okinawa Institute of Science and Technology (OIST)
Featured image: Artist impression of the transport proteins of SAR11 bacteria Credit: Kaori Serakaki | OIST